twisters
Newbie

At first, I did transient post-simulation for memory cicuit using finesim and extracting circuit using calibre xrc.
Using this method, I couldn't find out the nearest and furthest cell delay difference.
(Only the total parasitic value of the cell view taken as instance in the test bench was shown in the transient response)
My question are
1. Any of you who also checking on this delay difference? Which extraction and simulation tools are you using?
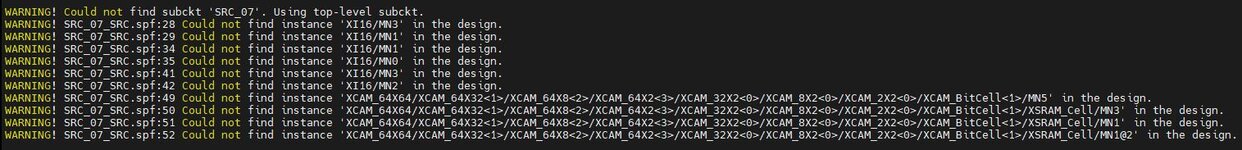
2. From my current observation, using the calibre xrc only produce net matching and failed for node matching.
While, the using SPF file from StarRC extraction can produce node matching but the matching rate is less than 1%.
Anyone know how to make the nodes matched?
Please refer to the attached image for the warning for unmatched nodes.
Using this method, I couldn't find out the nearest and furthest cell delay difference.
(Only the total parasitic value of the cell view taken as instance in the test bench was shown in the transient response)
My question are
1. Any of you who also checking on this delay difference? Which extraction and simulation tools are you using?
2. From my current observation, using the calibre xrc only produce net matching and failed for node matching.
While, the using SPF file from StarRC extraction can produce node matching but the matching rate is less than 1%.
Anyone know how to make the nodes matched?
Please refer to the attached image for the warning for unmatched nodes.