shaikh105
Member level 1

rdsamp.exe
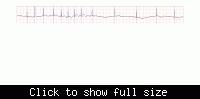
i have collected samples of ECG , 10 sec each. i converted them to one-D signal . Now the ECG signal is present with me as Ft=[a s d .......],,, ok
I want to remove
1. 60HZ frequency component
2. base line drift.
Can any body who played with ECG signal help me with some ideas using matlab.
i have collected samples of ECG , 10 sec each. i converted them to one-D signal . Now the ECG signal is present with me as Ft=[a s d .......],,, ok
I want to remove
1. 60HZ frequency component
2. base line drift.
Can any body who played with ECG signal help me with some ideas using matlab.